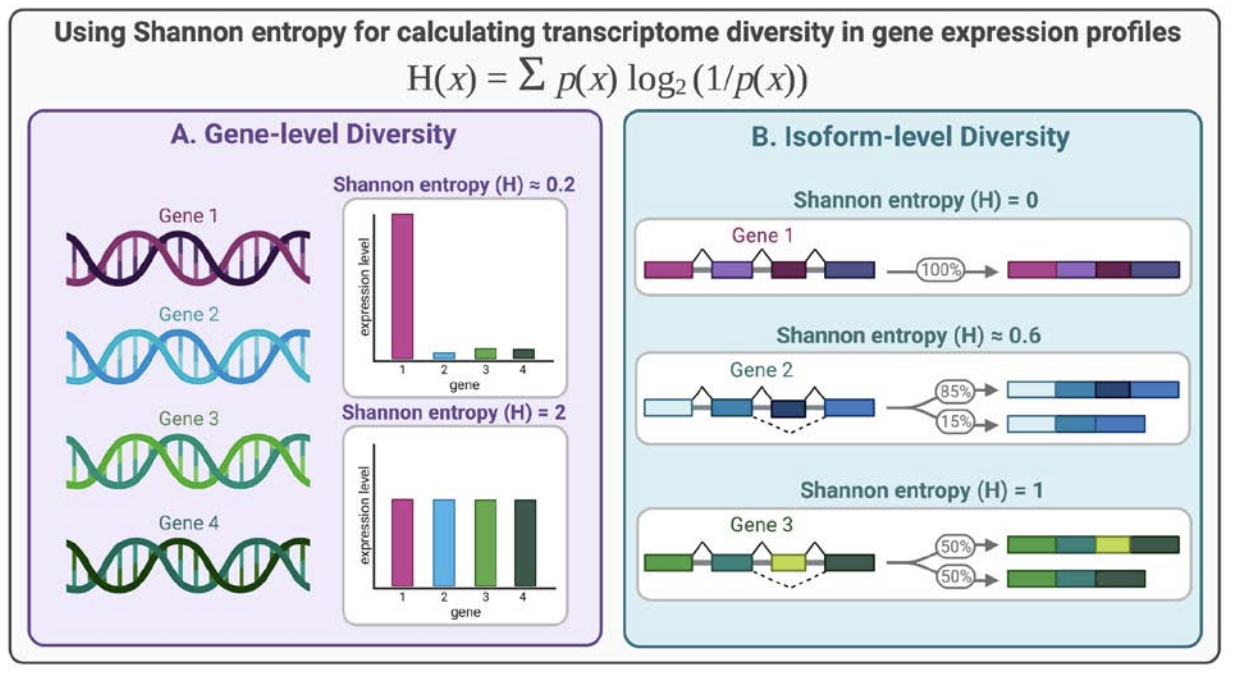
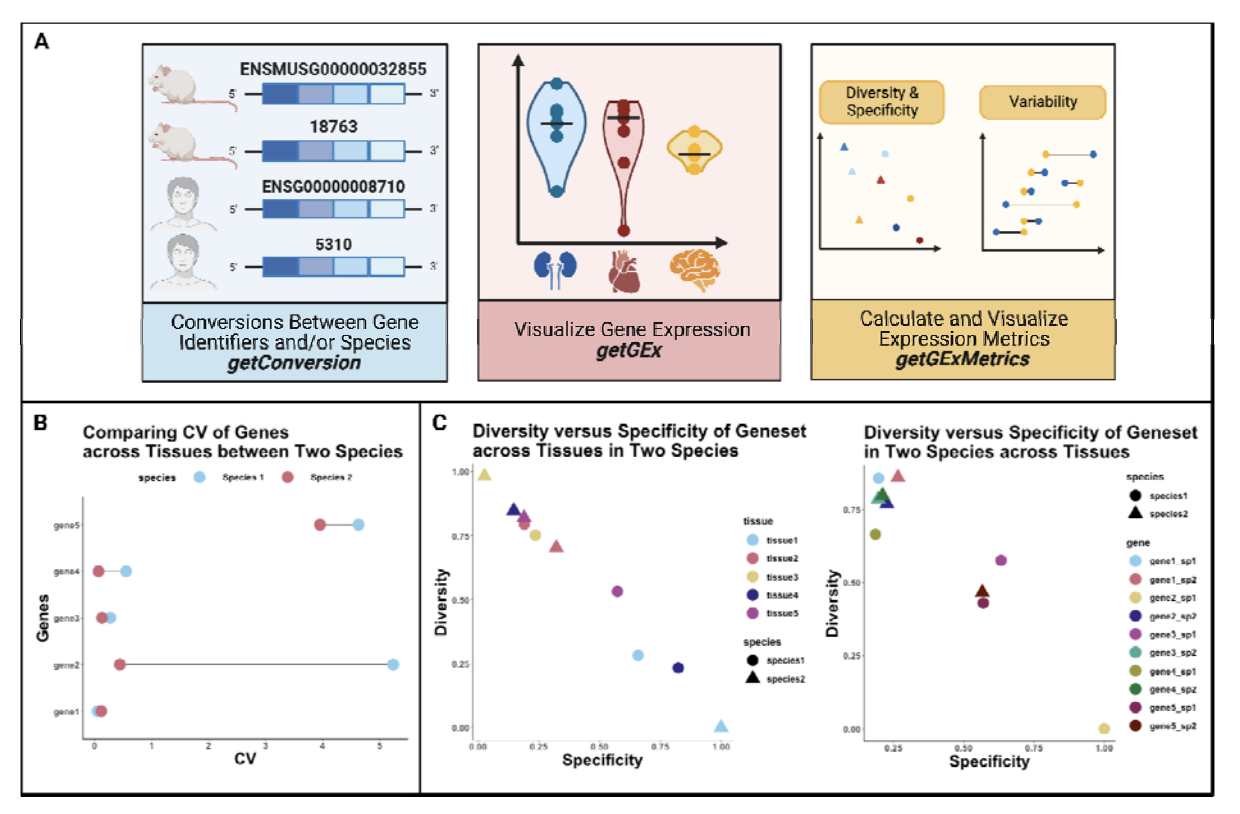
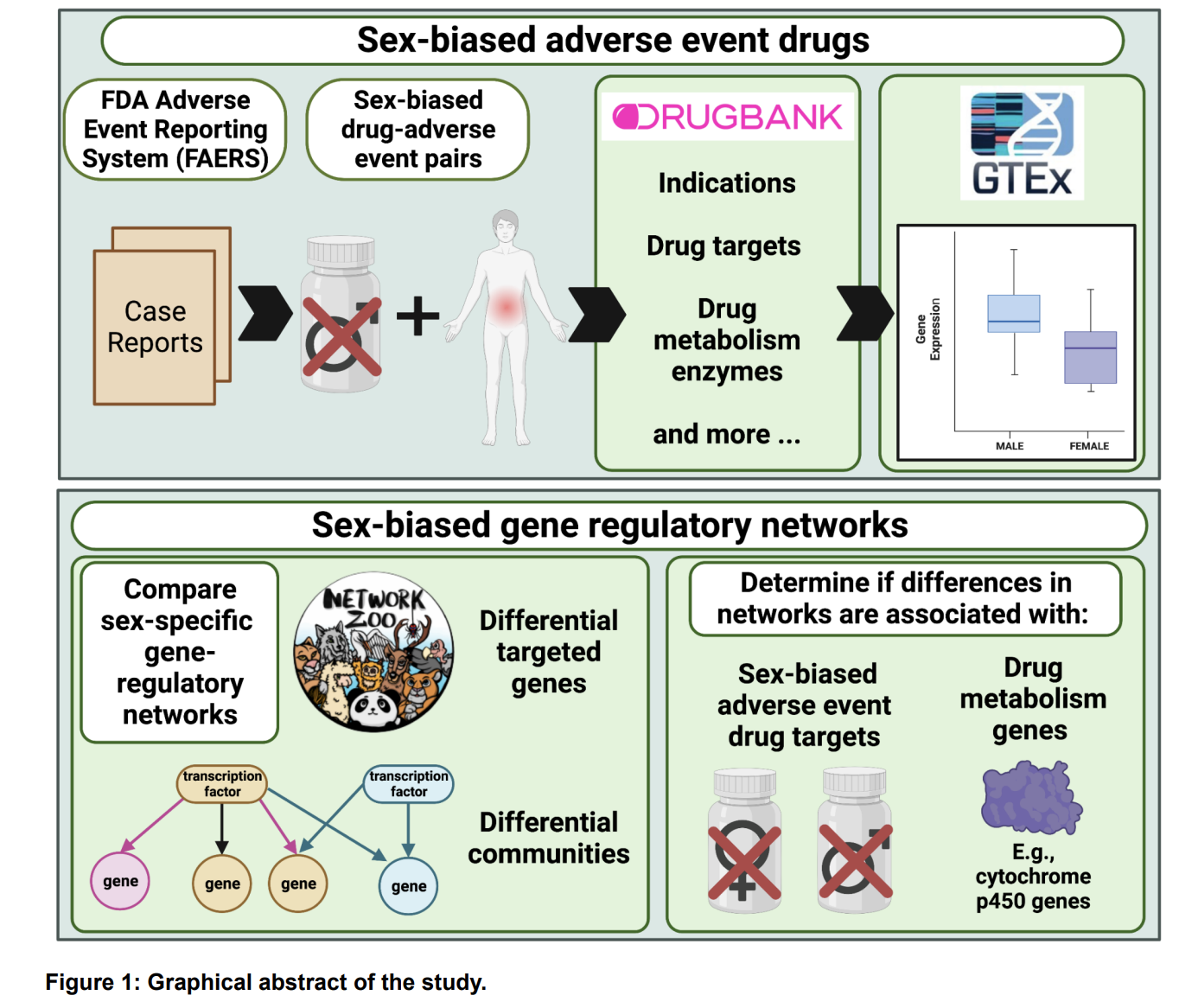
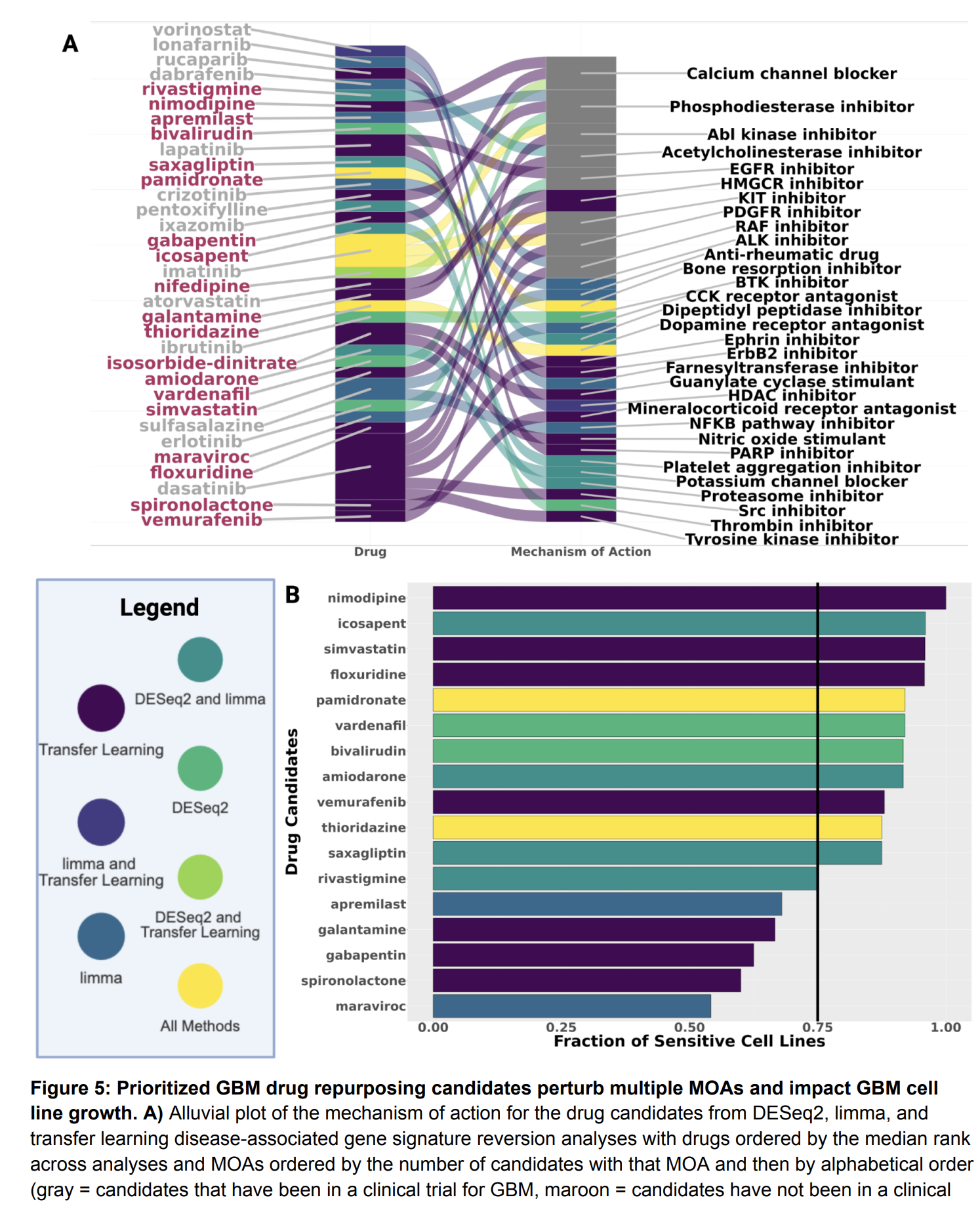
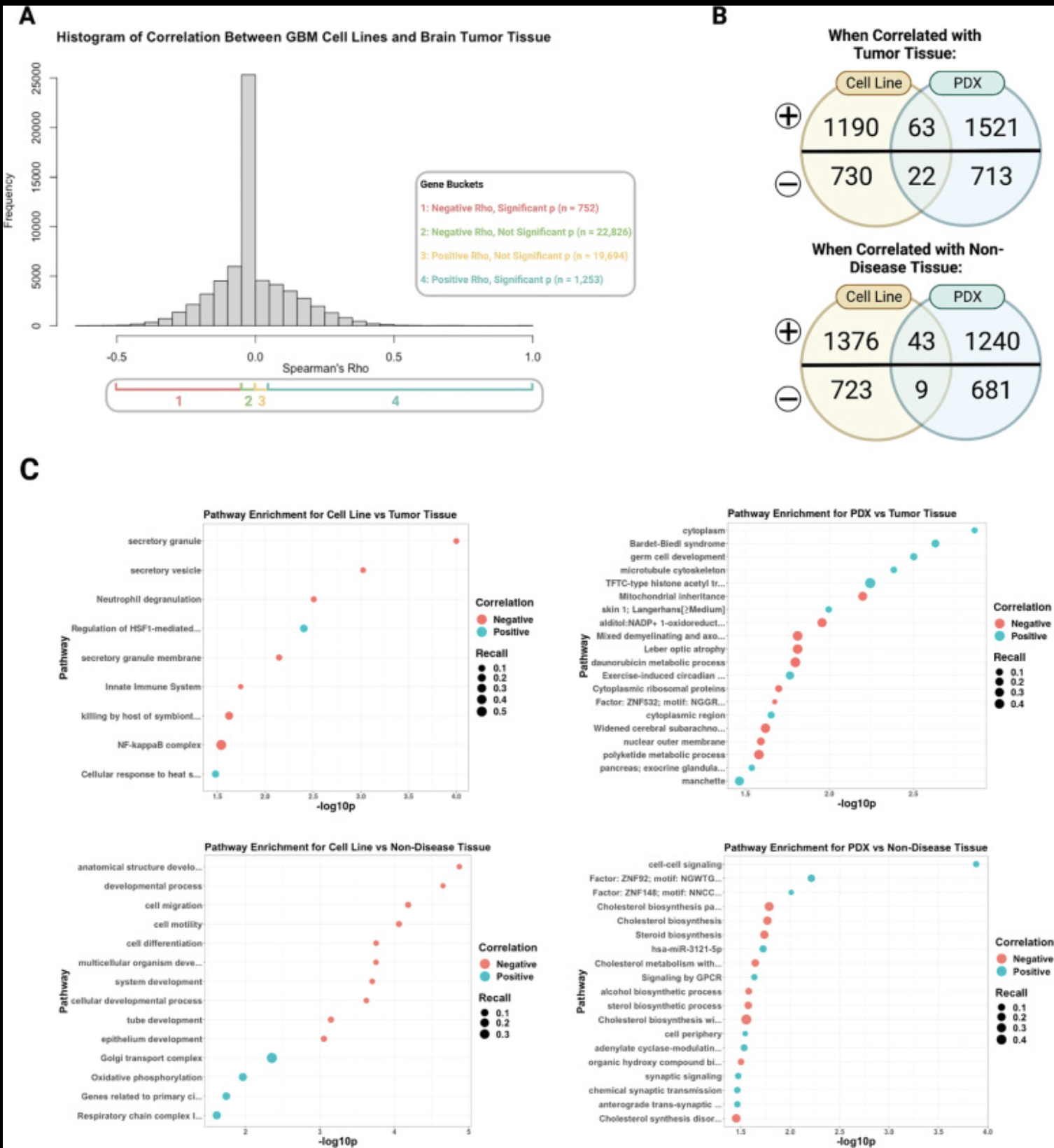
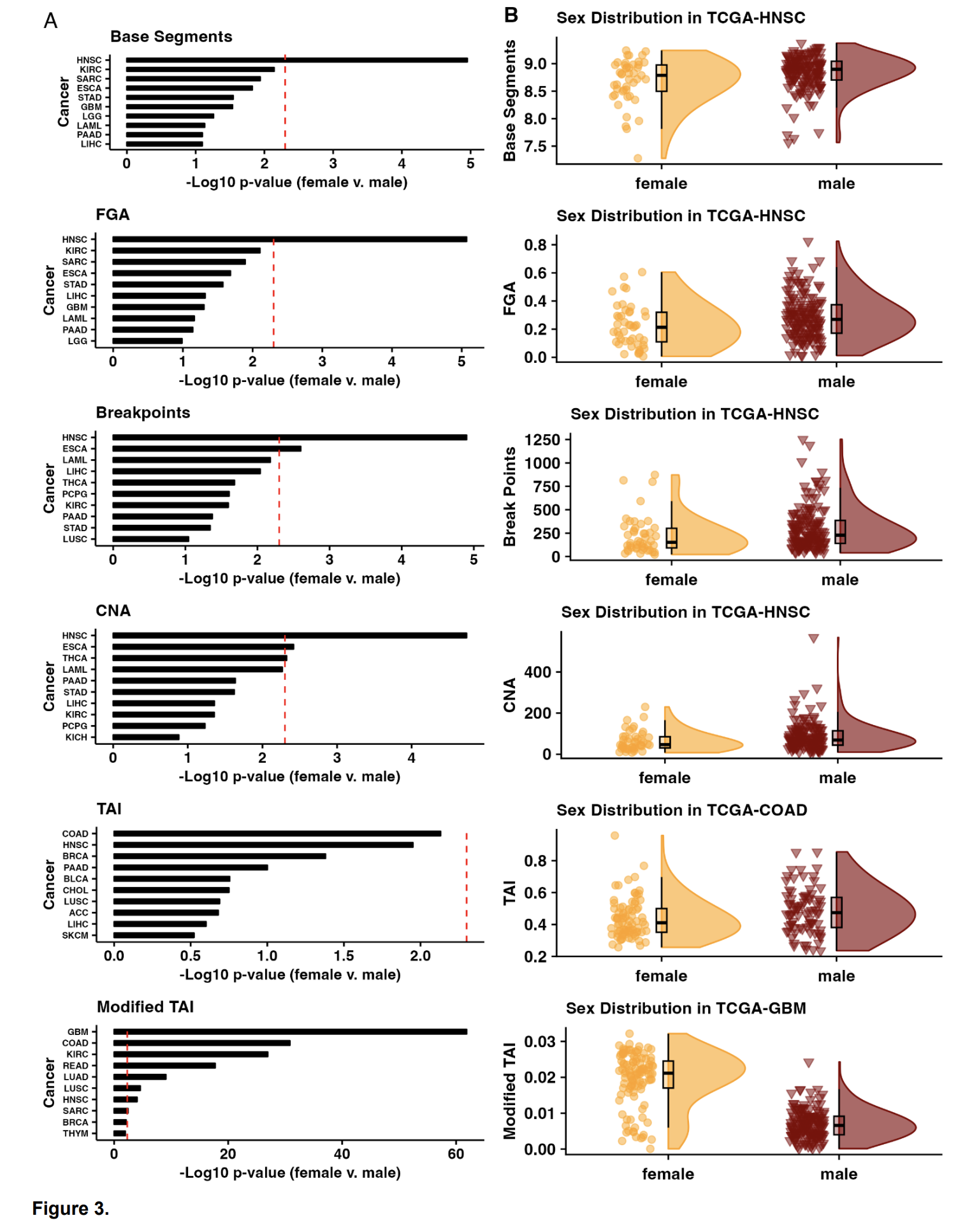
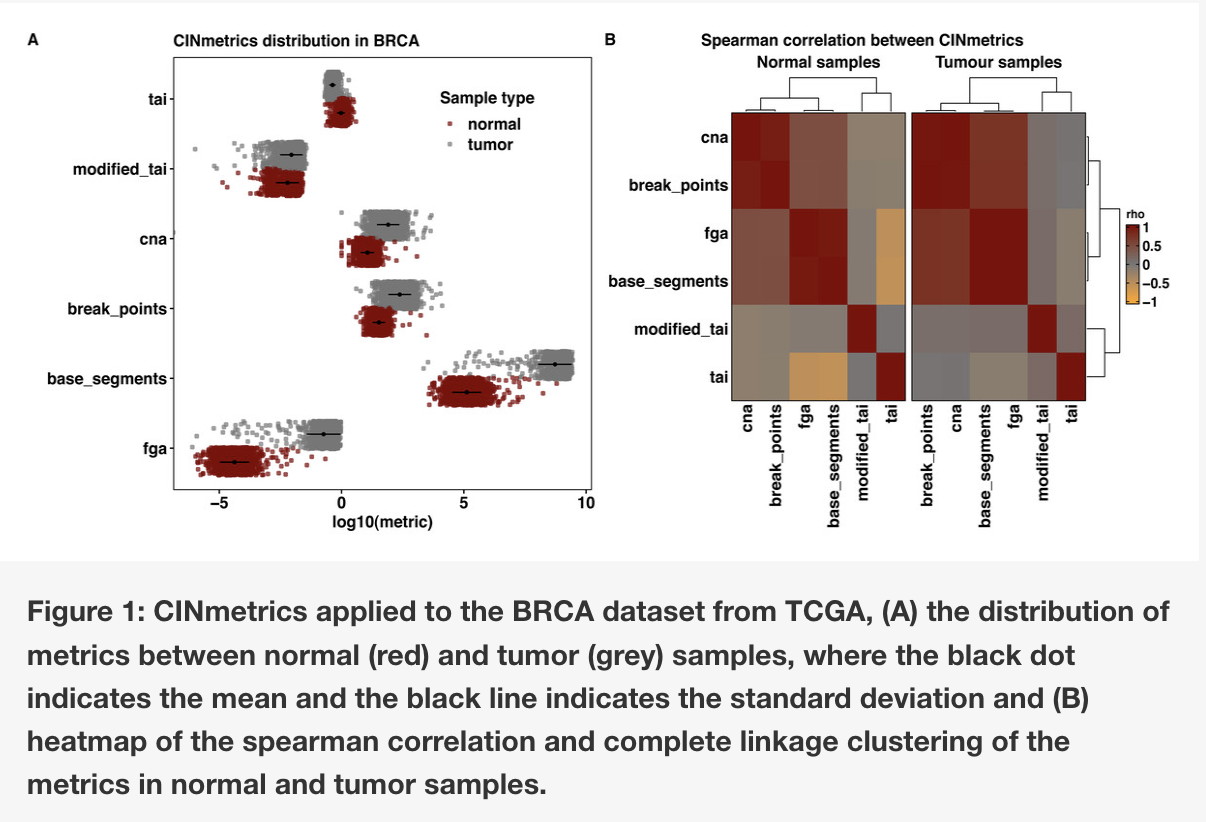
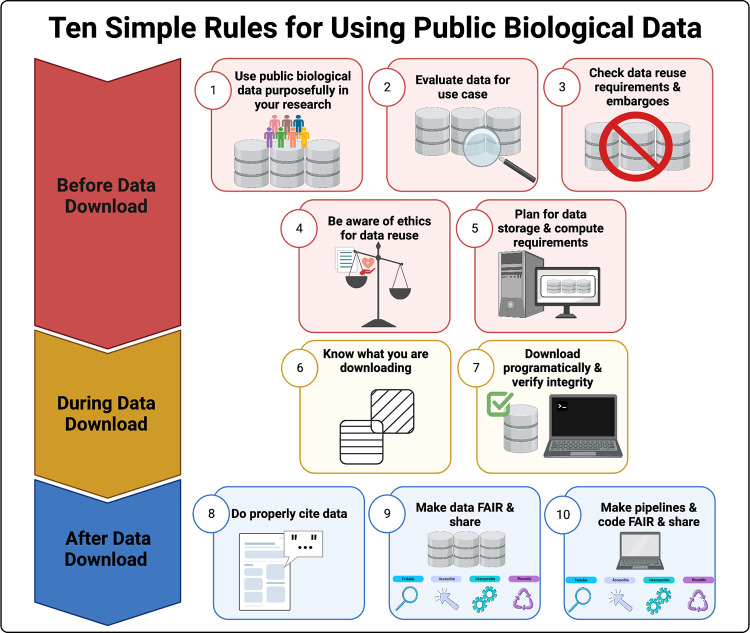
Objective/Rationale: This one day workshop is intended to provide HudsonAlpha undergraduate student BioTrain interns and graduate rotation students an introduction to computational biology through a discussion on why computers are necessary in genomics, short unix shell and R tutorials followed by hands-on exercises (The Carpentries lessons), and tips on how to ask for help, keeping a computational lab notebook, and finding additional learning resources.
Pre-requisites: Curiosity about computational biology and a basic understanding of the concepts of files and directories. Additionally, access to a laptop with the necessary software and data (see below).
8am-9am: Computer set-up This can be done anytime before bootcamp begins, but helpers will be available during this hour.
Please ensure you have the following software and data on the computer you intend to use during the workshop:
Make sure you have access to a Bash Shell (use Git BASH for Windows); and download the data-shell.zip file
Install a) R, b) RStudio, and c) download the r-novice-inflammation data
9am-10am: An Introduction to Biology with Computers (slides)
10am-12pm: Introduction to the unix shell
12pm-12:30pm: Keeping a Computational Lab Notebook & How to Get Programming Help (slides)
12:30pm-1:30pm: Lunch
1:30pm-4:00pm: Programming with R
4pm-4:30pm: What Now? (Continued Learning)(slides)
Check these shell lessons out if you:
-want to combine existing shell commands
-perform the same action on many files in the shell
-need to save and reuse shell commands
Check these R lessons out if you:
-want to practice more R programming with the inflammation data set used during bootcamp
-are interested in R programming lessons for reproducible scientific analysis